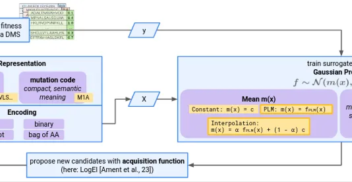
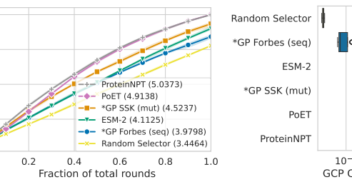
From our beginnings to becoming a key player in AI, supporting new research spaces and sharing what we’ve learnt in AI has always been at the core of InstaDeep’s DNA. This...

InstaDeep showcases 8 papers at NeurIPS 2024...
on Dec 09, 2024 | 03:16pm
InstaDeep delivers AI-powered decision-making systems for the Enterprise. With expertise in both machine intelligence research and concrete business deployments, we provide a competitive advantage to our customers in an AI-first world.
Learn More